Our Recent Work
Discover the latest work from our doctoral researchers! In this section, our students present their publications along with visuals that highlight their findings. For a full list of publications, visit the Publications tab in the main menu.
Protein recruitment
Ainara Claveras
Research Group: Gerhard Hummer
Institution: Max Planck Institute of Biophysics

To maintain cellular health, our cells get rid of damaged materials, such as protein aggregates, through a process known as autophagy. Thiscomplex process involves a double-membrane organelle called the autophagosome, which engulfs cellular debris and delivers it to the lysosome for degradation. If the autophagy machinery malfunctions, damaged proteins and organelles can accumulate in the cytosol and harm the cell. There is increasing evidence suggesting that defects in autophagy may be linked to neurological diseases like Parkinson’s. One of the first steps of autophagy is to recruit essential proteins to a specific site. The class III phosphatidylinositol (PI) 3-kinase complex 1 (PI3KC3-C1) plays a very important role in this process, as it phosphorylates lipids on the membrane of the autophagosome precursor. These phosphorylated lipids are then recognized by other autophagy proteins. Using a combination of cryo-EM and molecular dynamics simulations, we have obtained valuable insights into the mechanism of activation of the PI3KC3-C1 complex. Our data suggests that the catalytic subunit of the complex needs to reorient before binding to the membrane. Once this subunit engages with the membrane, lipids can access the catalytic site and undergo phosphorylation. We believe our findings will set the stage for future research into the subsequent steps of the autophagy pathway.
Defense against HIV
Jan Philipp Kreysing
Research Group: Martin Beck
Institution: Max Planck Institute of Biophysics
Publication: doi.org/10.1016/j.cell.2024.12.008
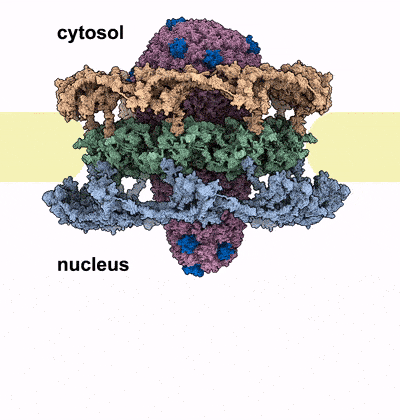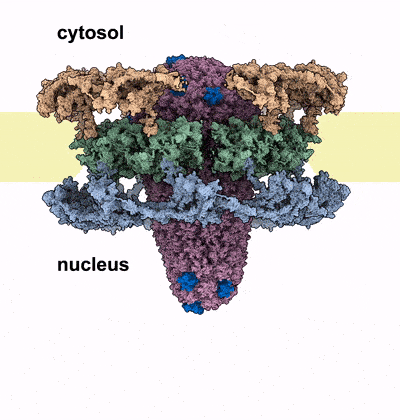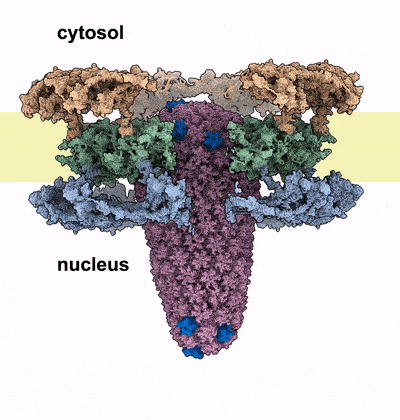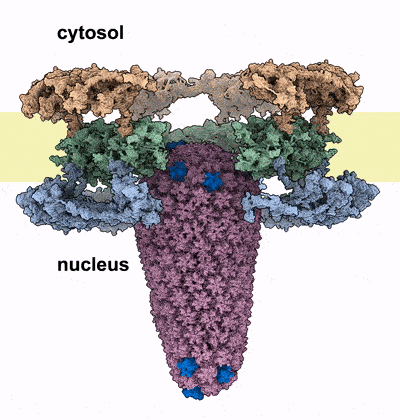
HIV-1 infection relies on the ability of the virus to transport its genetic material into the nucleus of human cells, where it integrates irreversibly into the host genome. Our work sheds light on the unique mechanism by which the HIV-1 capsid traverses the nuclear pore complex (NPC), a selective barrier guarding the nucleus. Using electron microscopy and computational simulations, we found that the cone-shaped capsid enters the nuclear pore with its narrow end first, exerting mechanical force that cracks the NPC’s ring-shaped structure, allowing the capsid to pass intact into the nucleus. This study further cements the critical role of HIV capsid in the HIV infection process, and introduces the novel concept of nuclear pore complex cracking. Better understanding of how the capsid helps the virus infect our immune cells is vital to developing better drugs against it.
Membrane Remodeling
Borna Markusic
Research Group: Ivan Đikić
Institution: Goethe University Frankfurt
Publication: doi.org/10.1073/pnas.2408071121
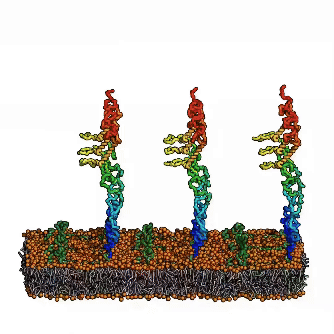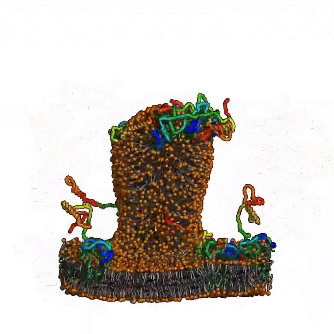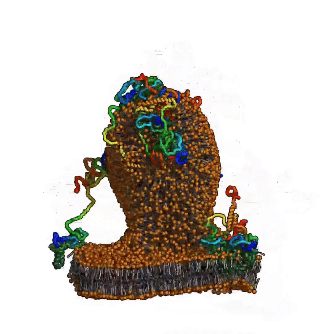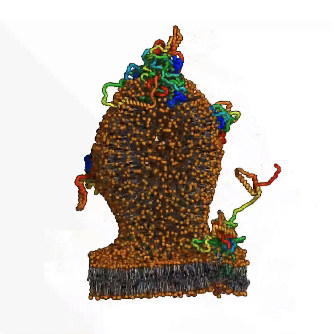
Until now, the involvement of intrinsic disorder in membrane shaping processes has not been fully understood. A multidisciplinary team around Ramachandra M Bhaskara from Institute of Biochemistry II (IBC2) led by Sergio Alejandro Poveda Cuevas now reports in PNAS detailed insights into how intrinsically disordered regions (IDRs) of membrane proteins remodel cellular membranes. Focusing on the ER-phagy receptor FAM134B, they explored how membrane-anchored disordered regions behave. Through advanced computer modeling and molecular dynamics (MD) simulations, the team found that – depending on context – these highly flexible protein regions exhibit different behaviors: Driven by their conformational entropy alone, they can sense and induce membrane curvature, thereby aiding in local remodeling. However, when combined with membrane-shaping elements like the Reticulon homology domain (RHD), they amplify large-scale remodeling processes by active scaffolding. This Janus-like behavior is sequence-encoded and shared among other proteins involved in ER-phagy. It allows IDRs to boost protein clustering and accelerate the reshaping of the ER, providing a fresh perspective on their role in regulation of membrane dynamics and shaping of cellular organelles.
Structure of NADPH oxidases
Victor Dubach
Research Group: Bonnie Murphy
Institution: Max Planck Institute of Biophysics
Publication: doi.org/10.1038/s41594-024-01348-w
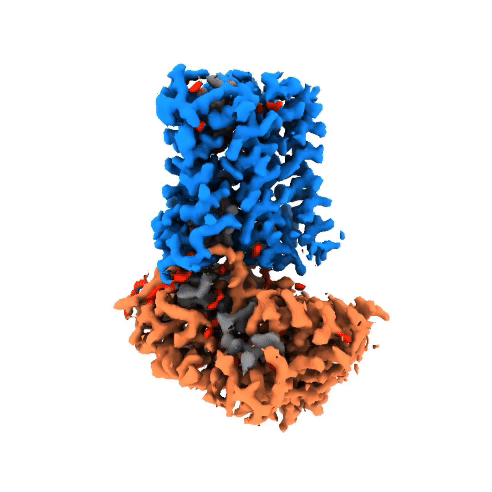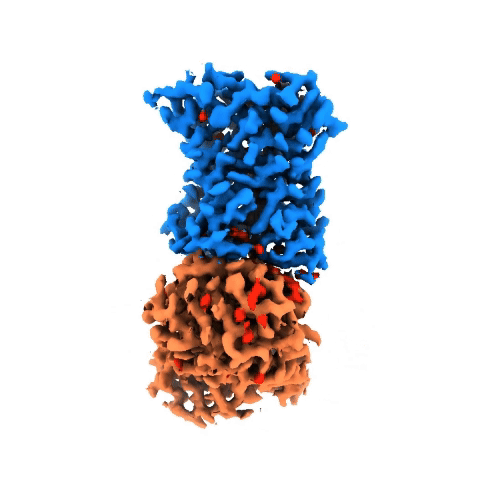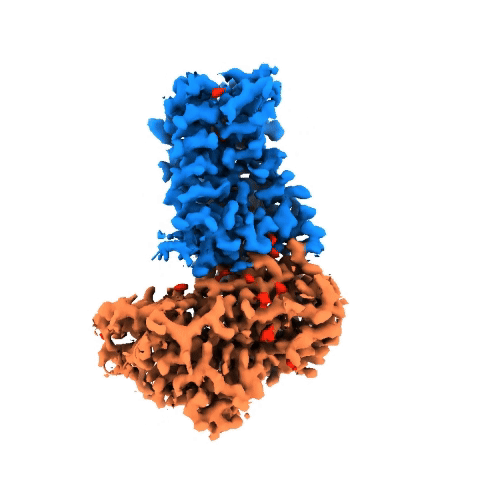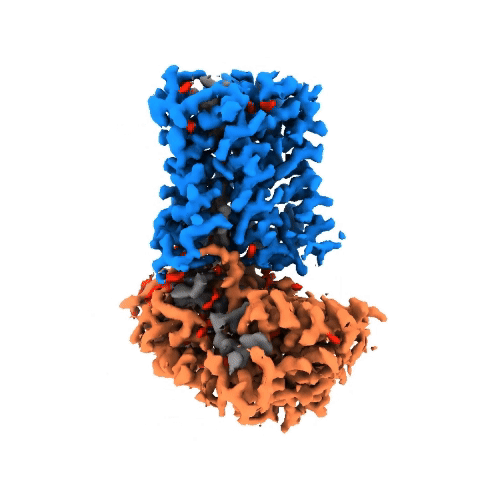
The NADPH oxidase (NOX) protein family plays an important role in the innate immune system, cell differenation and cancer. The enzyme oxidizes NADPH and use it as an electron source to reduce oxygen on the other side of the membrane. Even though NOXs have been studied for decades, there is little structural and mechanistic information on them. In order to gain insight, we have characterized a bacterial NOX homolog from Streptococcus pneumoniae (SpNOX). In total, four cryo-EM structures were obtained of the 46-kDa SpNOX. These structures were supplemented with targeted mutagenesis based on the obtained structures as well as (an)aerobic activity assays. These data gave insight into the lack of substrate selective of SpNOX opposed to its human homologous as well as the electron pathway across the membrane. Furthermore, the mechanism of the hydride transfer was elucidated allowing us to propose a catalytic cycle for SpNOX and other NOXs.



